Visium HD Spatial Transcriptomics Service
High-resolution spatial gene-expression profiling — we offer a standard workflow and optional extended post-processing.
Standard Workflow
BCL → FASTQ
spaceranger mkfastq demultiplexing
Primary Mapping
spaceranger count maps reads to genes and barcoded bins
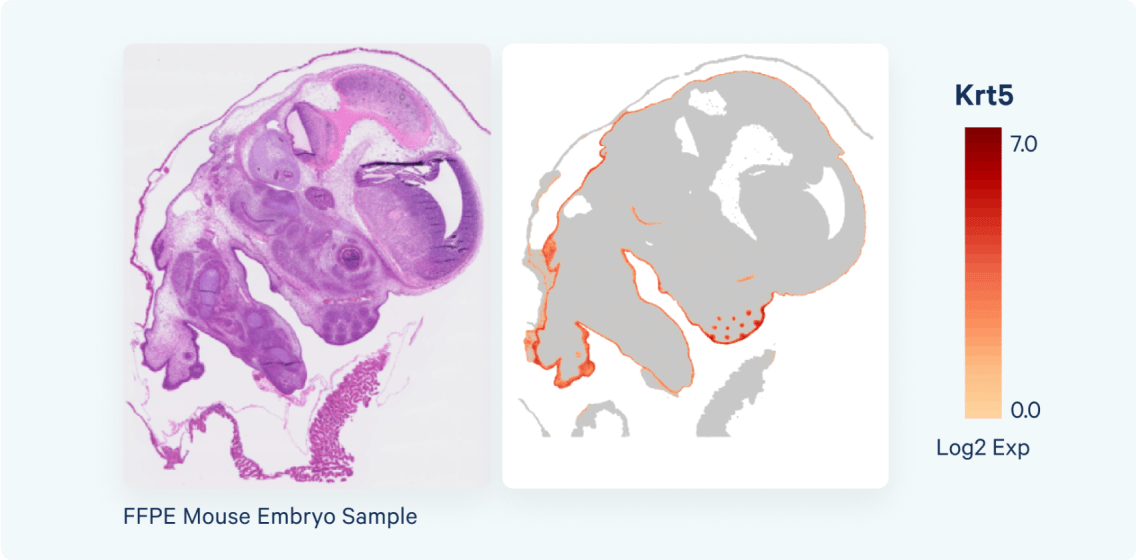
- FASTQ generation & Space Ranger run at 2 µm barcoded squares
- Interactive exploration at 8 µm resolution via Loupe Browser
- Comprehensive QC report (reads, UMIs, genes, tissue coverage)
- Delivery of raw & processed matrices (HDF5 / MTX)
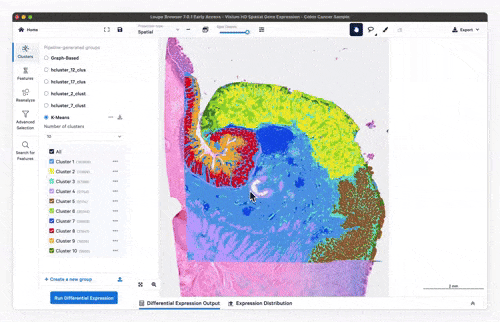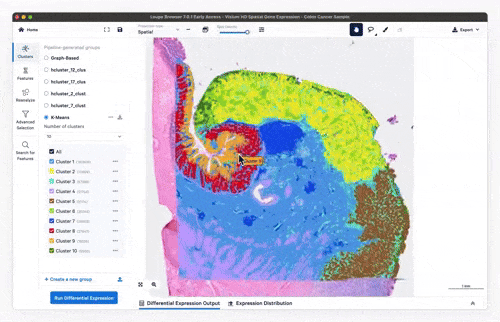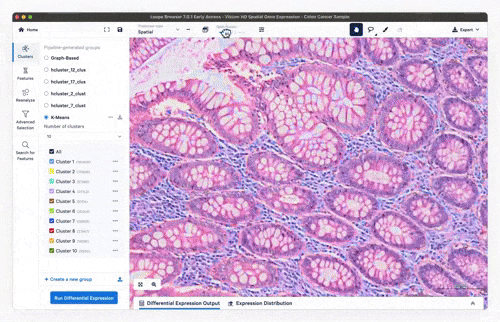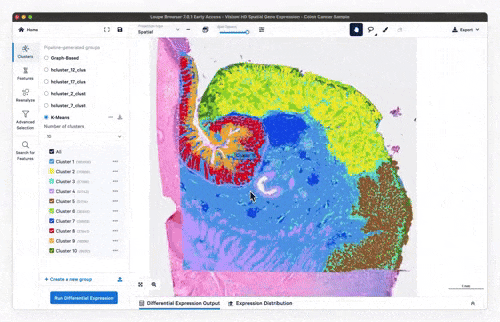
- Feature exploration, clustering & differential expression
Feature exploration
Clustering
Differential expression
Post-Processing (optional)
Choose from advanced spatial analyses to gain deeper biological insight.

Segmentation
StarDist / Bin2cell for cell-level matrices & masks

Neighborhood Enrichment
Squidpy analysis of cell–cell proximities

Spatially Variable Genes
Identify & rank genes with spatial patterns

Cell-Type Deconvolution
cell2location mapping using scRNA-seq references
Need something else? We’re happy to discuss custom pipelines.
What You Receive
Interactive Files
Loupe, H5Seurat / AnnData & HTML reports
Publication-Ready Figures
High-res UMAPs, heat-maps & overlays
Comprehensive QC
Tables & metrics for journals & grants
Ready to Get Started?
Contact us for a tailored quotation or consultation.

